Research areas
- Structural biology of protein-DNA complexes
- Phosphorylation mechanisms in Wnt signaling
- Small molecule inhibitors
- NMR methods
Main objectives
- Biochemical and structural characterization of protein-DNA interactions involved in DNA repair and transcription
- Design and pre-clinical evaluation of compounds for lead optimization
- Transforming biomolecular NMR to stay at the forefront of Structural Biology
Content of research
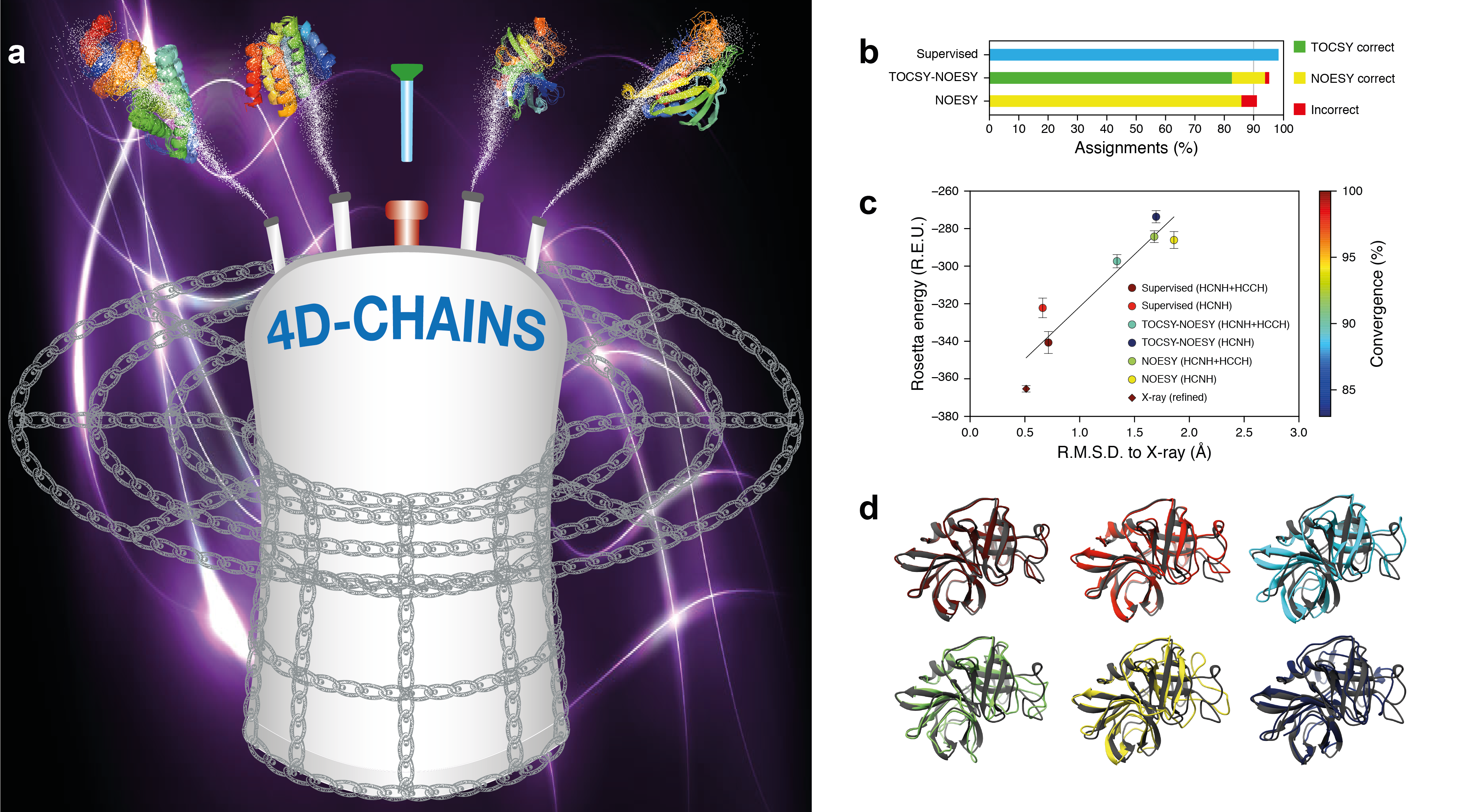
Ultrafast methods for NMR structure determination
The three-dimensional structure of a protein provides crucial insights into its biological function. NMR is the method of choice for proteins that are difficult, if not impossible, to crystallize or are relatively small (<50 kDa) for cryogenic electron microscopy to obtain atomic resolution. Yet, NMR data analysis is a laborious task and the road to NMR structure is generally a long one. The chief barrier is that existing methodologies require a significant amount of spectrometer time (several weeks), and effort by a trained expert (up to several months). We combine 4D NMR Spectroscopy with artificial intelligence to meet the key objectives of NMR structure determination; minimal data collection, least human intervention, and applicability to large proteins. We have developed 4D-CHAINS technology that utilizes only two 4D spectra and offers several advantages: (i) increased predictive power and higher reliability in sequential mapping by using all available correlated aliphatic chemical shifts, (ii) fully automated assignments of NMR chemical shifts at >95% completeness with <1.5% error rate, (iii) tremendous reduction in human effort and NMR spectrometer time needed to obtain data-driven, high-resolution structures, as the NOESY spectrum contains a set of distance constraints for structure calculations.
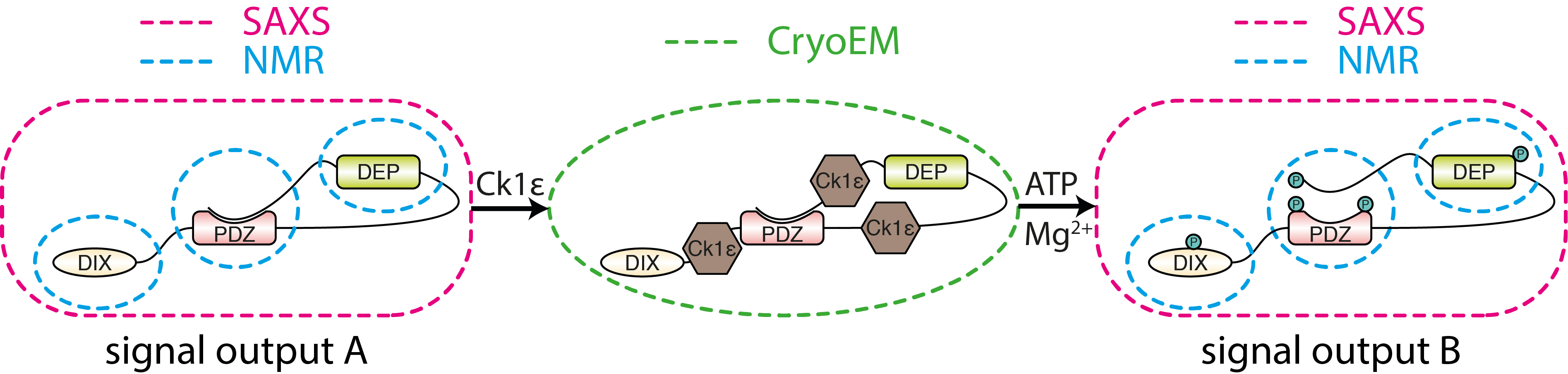
Mechanisms of protein phosphorylation
Phosphorylation, mediated by protein kinases, is a common posttranslational modification with 500,000 potential phosphorylation sites in the human proteome and 25,000 phosphorylation events described for 7,000 human proteins. Casein kinases are critical regulators of multiple developmental signaling pathways such as Wnt/β-catenin, Wnt/planar cell polarity, or Hedgehog. We use a suite of biochemical and structural biology tools in order to gain a mechanistic view of CK1ε action in the Wnt signaling pathways. Dishevelled (DVL) is a key target of CK1ε in the Wnt pathways, yet the mechanistic basis of DVL phosphorylation-driven activation is not understood. We apply integrated structural biology to (i) probe the DVL conformational landscape using in vitro and in vivo FRET sensors coupled to SAXS and CryoEM, (ii) understand the (auto)phosphorylation regulatory mechanisms of CK1ε, (iii) analyse by NMR the functional consequences of DVL phosphorylation and (iv) monitor DVL phosphorylation by real-time NMR under controlled cellular conditions.